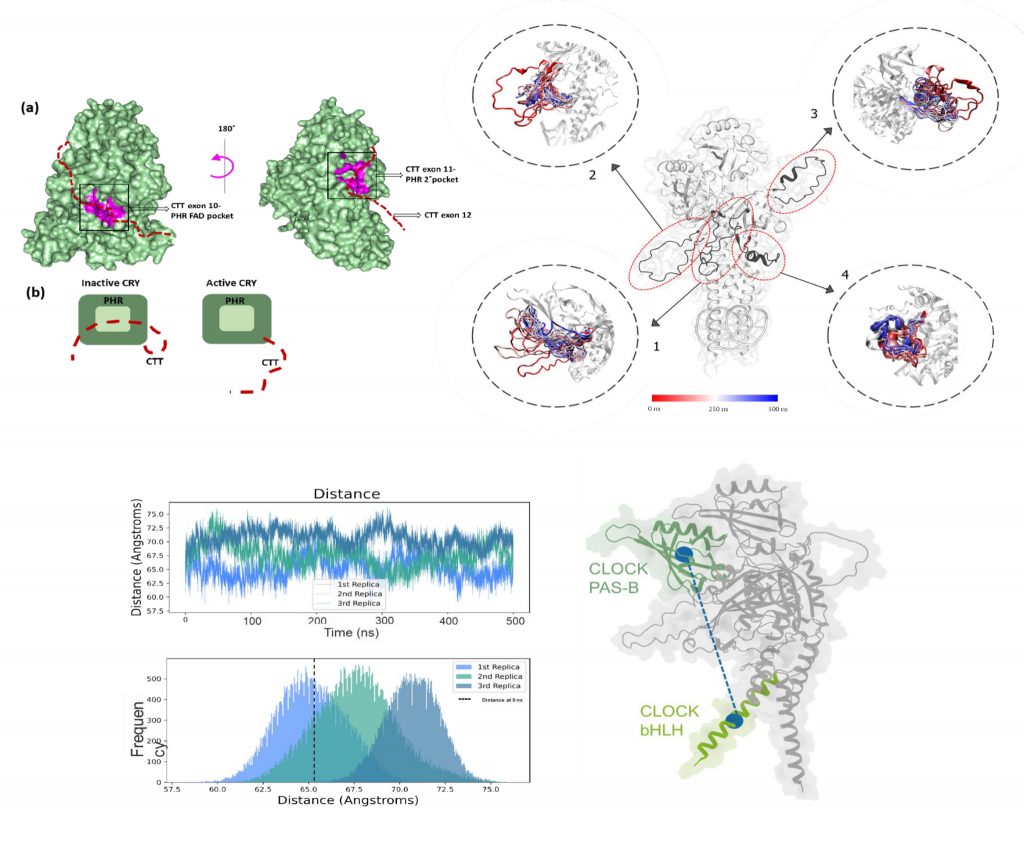
DYNAMICS OF BIOMACROMOLECULES
- Molecular dynamics simulations of clock proteins and lectins.
- Modeling intrinsically disordered regions.
- Understanding conformational changes in macromolecules.
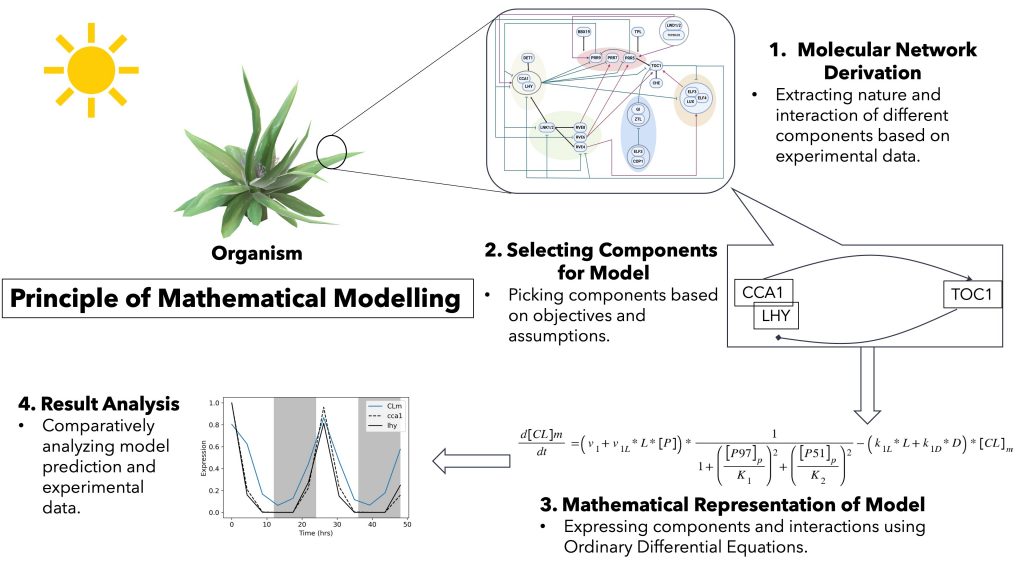
MATHEMATICAL MODELING
- Modeling different gene network circuits using Ordinary Differential Equations (ODEs).
- Understanding plant circadian rhythms under different physiological conditions.
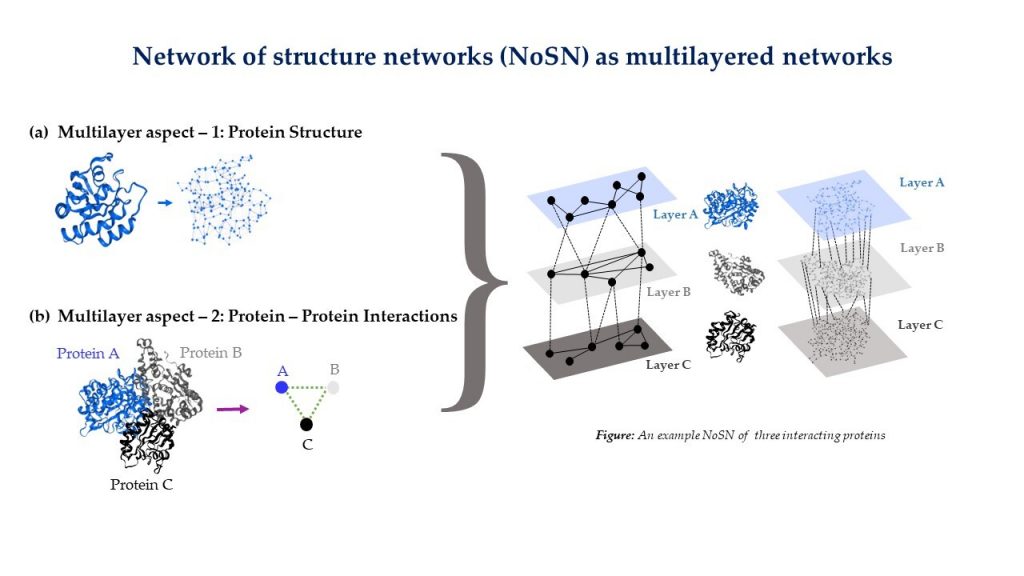
NETWORK BIOLOGY
- Network biology approach to understand protein structures and function
- Reconstruction of gene regulatory networks in circadian skeletal-muscle transcription.
- Multilayer modeling of Network of Structure Networks.
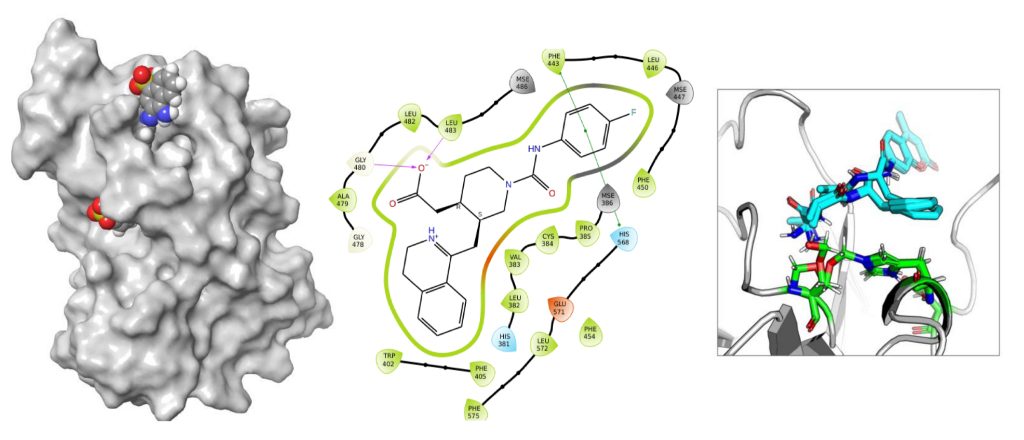
DRUG DISCOVERY
- Structure based virtual screening of compounds.
- To explore the transient or cryptic novel binding pockets.
- To predict novel compounds that bind to core clock proteins.
- Studying the dynamics of protein – ligand complexes.
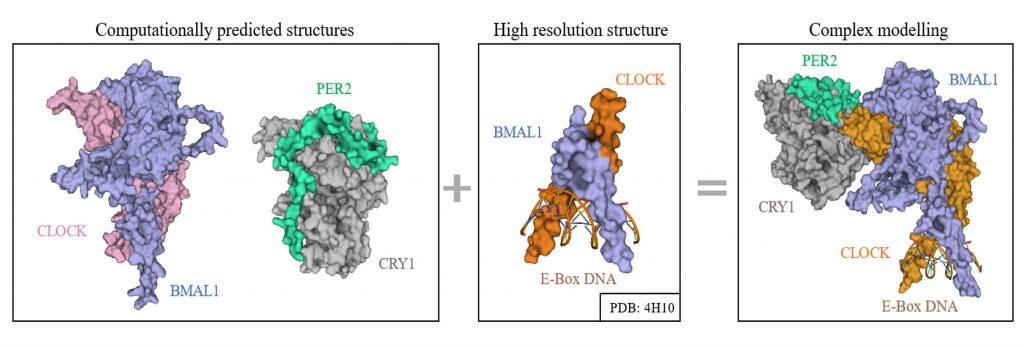
INTEGRATIVE MODELING
- Using computational methods to combine data from experiments like cryo EM, NMR, XRD, SAXS and XFEL.
- To explore structure & dynamics of circadian complexes.